Results in IMAT refer to data that is output from a process. This could be output from a finite element solution from I-deas, or it could be results from a modal test. One specific subset of results can be stored in IMAT as an imat_shp. In addition to the actual data values, results also have many attributes that help describe the nature of this data. Attributes include ID Lines, analysis source, result type (acceleration, stress, etc.), and many others. IMAT keeps all of these attributes and the data together in a convenient result object data type.
IMAT+FEA utilizes the imat_result data type for result data imported from NASTRAN and Abaqus files. In these cases not all of the attributes available in the imat_result object are applicable, but using this data type offers consistency within MATLAB.
Before discussing how to use the imat_result class (data type), it may be helpful to describe how it is organized.
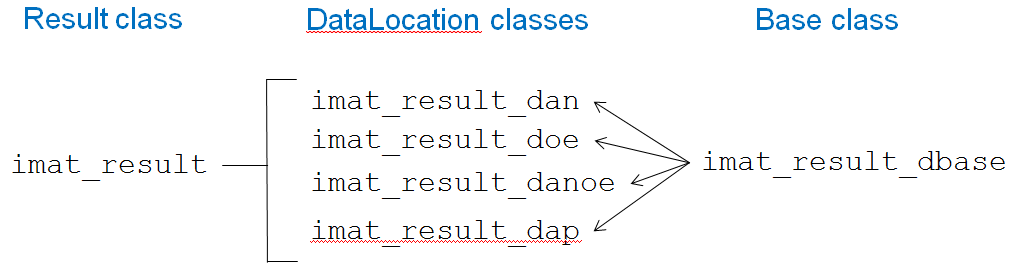
The imat_result class actually consists of a hierarchy of classes. Each individual result is defined in a class specific to the type of data of which it consists. All of the individual results are gathered in the top-level imat_result class, which contains most of the descriptive attributes. The class hierarchy is shown in the figure below.

One powerful feature of this class organization is that it is possible for the user to create their own result data location class and have it automatically included in the imat_result hierarchy. It only needs to be a subclass of imat_result_dbase, and define the methods and properties that the other result location classes define.
The table below describes all of the imat_result classes and their relationships.
| imat_result_dbase | This is the base class that describes the properties and methods (including abstract) for each of the result location classes. |
| imat_result_dan | Defines the properties and methods for a single Data At Nodes result. Subclass of imat_result_dbase. |
| imat_result_doe | Defines the properties and methods for a single Data On Elements result. Subclass of imat_result_dbase. |
| imat_result_dap | Defines the properties and methods for a single Data At Points result. Subclass of imat_result_dbase. |
| imat_result_danoe | Defines the properties and methods for a single Data At Nodes On Elements result. Subclass of imat_result_dbase. |
| imat_result | Defines the properties and methods for the overall results. It defines the descriptive attributes common to all of the results. It can be a vector or matrix of results. Each individual result is one of the imat_result_dbase-derived classes above. |
Assuming you have already worked with shapes in IMAT, you will find that results are treated in a similar way. An imat_result variable contains both the result data and various numeric and string attributes.
Results can be generated for various locations. Results can be data at a specific node, data on each of the nodes of an element (data at nodes on elements), data on an element, and others. Corresponding to the data locations and the data are data components which specify where the data values are located. For data at nodes, the component has three parts: the node number and the degree of freedom (component) on that node, such as X+, and the superelement ID. For data at nodes on elements, the component includes the element number, component (such as XX), and the node number on that element, the layer ID, superelement ID, and element type.
The imat_result object has properties for the attributes, the data (split into the various types of data), and other supporting attributes. To see the available properties and their expected data types or valid values, you can use the set function:
>> set(imat_result)
Name: [ String ]
IDLine1: [ String ]
IDLine2: [ String ]
IDLine3: [ String ]
IDLine4: [ String ]
IDLine5:
[ String ]
ModelType: [ Unknown | Structural | Heat transfer | Fluid flow ]
AnalysisType: [ Unknown | Static | Normal mode | Complex eigenvalue first order
| Transient | Frequency response | Buckling
| Complex eigenvalue second order | Static non-linear Craig-Bampton constraint modes
| Equivalent attachment modes | Effective mass modes | Effective mass matrix 1
| Effective mass matrix 2 | Distributed Load Load Distribution | Distributed Load Attachment modes ]
ResultType: [ Stress | Strain | Element Force | Temperature | Heat Flux
| Strain Energy | Displacement | Reaction Force | Kinetic Energy | Velocity
ExpLength: [ Numeric value ]
ExpForce: [ Numeric value ]
ExpTemperature: [ Numeric value ]
ExpTime: [ Numeric value ]
...
Time: [ Numeric value ]
Frequency: [ Numeric value ]
Eigenvalue: [ Numeric value ]
EigenvalueReal: [ Numeric value ]
EigenvalueImag: [ Numeric value ]
ModalMass: [ Numeric value ]
ModalMassReal: [ Numeric value ]
ModalMassImag: [ Numeric value ]
ViscousDamping: [ Numeric value ]
HystereticDamping: [ Numeric value ]
ModalA: [ Numeric value ]
ModalAReal: [ Numeric value ]
ModalAImag: [ Numeric value ]
ModalB: [ Numeric value ]
ModalBReal: [ Numeric value ]
ModalBImag: [ Numeric value ]
Stiffness: [ Numeric value ]
StiffnessReal: [ Numeric value ]
StiffnessImag: [ Numeric value ]
EffectiveMass: [ 6x1 numeric vector ]
ParticipationFactor: [ 6x1 numeric vector ]
data: [ IMAT_RESULT_* object]
>>
The specific meaning of each attribute can be found in the reference section. In general, you will have access to the data coefficients, the component labels, and three types of attributes:
Each of the individual result DataLocation classes also has its own set of attributes, which are described in the reference section. You can see the list of attributes for each by using the MATLAB properties method, as shown below.
>> properties(imat_result_dan) % Data At Nodes
Properties for class imat_result_dan:
DataLocation
NumberNodes
NumberValues
DataCharacteristic
component
node
dir
seid
data
numDisplay
You will see later how to examine and modify the data values and data attributes.
Just as for functions and shapes, multiple results can be arrayed in a single imat_result variable. Most often, you will work with a "column vector" of results (number of results by 1). The various elements of a result array need not be compatible, but most operations work most effectively when they are.
If you can think of a use for multidimensional arrays of results, IMAT will happily work with them.
To create an imat_result from scratch, you use the imat_result constructor. To set the type of data stored in an individual result index, simply set the .data attribute to an object of the datatype you want to use. You can also pass this object into the constructor, as shown in the examples below.
>> r = imat_result % Create a scalar imat_result
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
>> r = imat_result(3) % Create a 3x1 imat_result
r =
3x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
2 Data At Nodes Unknown User defined
3 Data At Nodes Unknown User defined
>> r = imat_result('ResultType','stress') % Change the ResultType
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown Stress
>> r.data = imat_result_danoe % Change the DataLocation (different from result object)
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------------------- --------- ------------
1 Data At Nodes On Elements Unknown Stress
>> r=imat_result('ResultType','stress',imat_result_danoe) % Set the DataLocation directly in the constructor (different from result object)
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------------------- --------- ------------
1 Data At Nodes On Elements Unknown Stress
After you create a result, you will want to set its attributes and data values. Click here to see how to do this.
It is also possible to convert an imat_fn or imat_shp into an imat_result, and vice versa. Simply pass the variable into the result constructor function as shown below.
>> f=imat_fn(3,'responsenode',1:3,'ordinate',reshape(1:18,6,3)); % Creates 3x1 imat_fn
>> r=imat_result(f); % Creates 6x1 imat_result (one for each abscissa value in f)
There are several ways to get results from I-deas and other software packages into MATLAB. Results are transferred through Universal files in dataset 2414. They are also returned from readnas and readodb for the appropriate data types.
The IMAT toolbox allows you to read a wide variety of universal files. Universal files are generally ASCII text files that can be read and written by I-deas and other software packages. You can create a universal file containing results by accessing the Export menu in I-deas.
You create a result universal file in I-deas using the File/Export menu selection. Select the option "Simulation Universal File" option, and be sure to highlight the results.
To read a results universal file into MATLAB, use the readunv function in the IMAT toolbox. You can provide a filename argument, or call readunv with a wildcard or no argument to interactively select the file to read. Note that readunvwill by default read mode shape results in as an imat_shp. To read them in as a result, use the 'asresult' optional input argument.
When transferring universal files between platforms, be sure to use an ASCII (text) file transfer protocol so that carriage returns get converted properly.
You have one option for transmitting results to I-deas and other software packages.
You can create a result universal file (dataset 2414) using the writeunv function in the IMAT toolbox. The first argument is a filename, which can be a wildcard. The remaining arguments are one or more imat_result variables to be written to the universal file. You can also combine these variables with the other IMAT objects.
You read the shape into I-deas using the File/Import menu selection using the "Test Universal File" or the "Simulation Universal File" option, depending on what application you are in.
If you copy the universal file between platforms, you should be sure to use an ASCII file transfer protocol so that carriage returns get converted properly.
IMAT offers two methods for you to examine or modify the contents of an imat_result.
The simplest way to get information about a result is with the syntax r.attrib, where r is the name of the variable, and attrib is the name of the attribute you want. The possible attribute names are listed in the reference section of this document, or you can get a listing with the set function. Here are some examples:
>> r.frequency
ans =
1.0
2.1
3.224
27.56
>> r.name
ans =
' B.C. 2,DISPLACEMENT_11,LOAD SET 1'
' B.C. 2,REACTION FORCE_12,LOAD SET 1'
' B.C. 2,STRESS_13,LOAD SET 1'
' B.C. 2,STRAIN_14,LOAD SET 1'
>>
An alternative way to access shape information is with get. In general, the expression get(r,'attrib') is equivalent to the expression r.attrib as discussed above. However, the get function also allows you to extract multiple attributes in one call. Here's an example:
>> g=get(r,'idline1','loadset','datalocation','resulttype')
g =
IDLine1: {4x1 cell}
LoadSet: [4x1 double]
DataLocation: {4x1 cell}
ResultType: {4x1 cell}
>>
When you ask for multiple attributes, get returns a structure with field names equal to the attributes you selected. The value in each field is the value of the corresponding attribute, as discussed above. If you do not specify any attributes at all, then get will return all of the data attributes. After a call to get, you can refer to the individual returned values by name, provided you use the proper combination of upper and lower case to refer to the fields:
>> r.ResultType
ans =
'Displacement'
'Reaction Force'
'Stress'
'Strain'
>>
These examples illustrate that numeric attributes like Frequency are returned in a numeric array the same size as r. String-valued attributes like Name as well as list attributes are returned either as a character string (if r is a single result or in a cell array of strings if r is a vector or array of results. (For more information on cell arrays and cell arrays of strings, consult the MATLAB documentation.)
You can access the components and data of a given imat_result through the .data property, which contains the result DataLocation object for a given imat_result. There are several ways to access the components. The imat_result class does allow you to access some of the underlying DataLocation result properties, but for complete access you will need to access the underlying objects directly.
To retrieve the entire component matrix directly, use the .component property, as shown below. However, you cannot set the component matrix this way.
>> r
r =
2x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- --------------------------------- ------------------------- ---------- ------------
1 SUBCASE=1, LOAD ID=1, TYPE=STRESS Data At Nodes On Elements Structural Stress
2 SUBCASE=2, LOAD ID=2, TYPE=STRESS Data At Nodes On Elements Structural Stress
>> r(1).data.component(1:10,:)
ans =
1 1 11 1 0 0
1 1 12 1 0 0
1 1 22 1 0 0
1 1 13 1 0 0
1 1 23 1 0 0
1 1 33 1 0 0
1 1 11 2 0 0
1 1 12 2 0 0
1 1 22 2 0 0
1 1 11 2 0 0
>> r(1).data.data(1:3)
ans =
1.23435
3.65766
0
It can be more useful to access the components by their individual columns. Each result DataLocation object has a different set of property names for the component columns, since each one has a different set of columns. Below are some examples, based on the result shown above. You can both get and set the component list this way. When setting these attributes, the object will automatically extend or truncate all of the component columns and the data if you change the number of values.
>> r(1).data.element(1:4)
ans =
1
1
1
1
>> r(1).data.comp(1:4)
ans =
11
12
22
13
Using the individual data component columns can be a very handy way of extracting a subset of results. For example, to extract node numbers 1 and 2 and direction X+ from a Data At Nodes result, you can use the following code snippet.
>> ind = ismember(r(1).data.node,[1 2]) & r(1).data.dir == 1; % Build a logical index vector that matches nodes 1 and 2, direction 1
>> r(1).data.component(ind,:) % Extract the component submatrix
ans =
1 1 0
2 1 0
>> r(1).data.data(ind) % Extract the corresponding data
ans =
1.1
1.2
Data attributes of an imat_result return a single numeric or string value for each element of a result array. The Component and Data "properties", on the other hand, each return an array of values for each element of the result array. When you ask for the .data.data attribute (data values for each of the results), you will get a multidimensional array whose first dimension corresponds to the number of data values, and whose remaining dimensions match the size of the imat_result array.
The same thing happens when you access the .data.component attribute (the data components): an extra dimension is added as the first dimension of the result.
If not all of the results have the same number of data values or components, IMAT sets the first dimension of the result to the largest of any of the results in the array. For those results with less data than the maximum, the extra elements are filled with NaN (i.e., "not a number") values.
Examples of this are shown below.
>> r
r =
2x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- --------------------------------- ------------------------- ---------- ------------
1 DERIVED RESULT Data On Elements At Nodes Structural User defined
2 SUBCASE=2, LOAD ID=2, TYPE=STRESS Data At Nodes On Elements Structural Stress
>> r.data.component
ans(:,:,1) =
1 1 1 0 NaN NaN
1 1 2 0 NaN NaN
1 1 3 0 NaN NaN
ans(:,:,2) =
1 1 11 0 0 0
1 1 12 0 0 0
1 1 22 0 0 0
>> r.data.data
ans =
1.23435 6.53175
3.65766 -3.65766
0 0
Sometimes it is useful to partition results to a subset of components. One way to achieve this is to index into the component matrixas shown in the examples above, and then use setComponents to build a new result. However, that can be cumbersome. IMAT provides partition methods to wrap this capability into an easy-to-use method. In addition, it allows you to use imat_ctrace, imat_group, and numeric submatrices, depending on the result DataLocation. Not all data types support all of these input types, and the way some of these partitioning inputs are interpreted vary with the result DataLocation.
In the following example, we will start with a scalar imat_result containing Data At Nodes data. We will then partition the imat_result_dan object in the .data property using an imat_ctrace and a numeric submatrix. Following that, we will show that you can also partition the result using the partition method on the imat_result itself. Please note that if your result contains multiple DataLocations, this method may fail because not all partitioning input types are recognized by each of the different DataLocations.
>> r
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
>> dan=r.data % Extract the Data At Nodes object
dan =
Data At Nodes: 3DOF global translation vector
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 2 0 2
1 3 0 3
11 1 0 4
11 2 0 5
11 3 0 6
------------------------------------------------
Number of Values: 6 Number of Nodes: 2
================================================
>> ct=imat_ctrace('1y','11z') % Create an imat_ctrace to use for partitioning
>> dan.partition(ct) % Partition at the imat_result.data level
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 2 0 2
11 3 0 6
------------------------------------------------
Number of Values: 2 Number of Nodes: 2
================================================
>> dan.partition({[1 2; 11 3]}) % Look for matches in the first 2 columns; notice that the submatrix is passed in as a cell containing the submatrix
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 2 0 2
11 3 0 6
------------------------------------------------
Number of Values: 2 Number of Nodes: 2
================================================
>> r2=r.partition(ct) % You can also partition at the imat_result level, but this only works if all of the DataLocations support the supplied partitioning input
r2 =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
>> r2.data
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 2 0 2
11 3 0 6
------------------------------------------------
Number of Values: 2 Number of Nodes: 2
================================================
The numeric submatrix method of partitioning has the additional advantage of being able to specify wildcards by specifying inf for a given component value. In the example below, we are specifying two different wildcard matches. In the first row, we are asking for all components whose direction is 2. In the second row, we are asking for all rows with a node number of 11. Notice that the two matches overlap, but only the unique match is returned since the component rows must be unique in a given DataLocation object.
>> dan.partition({[inf 2; 11 inf]}) % Look for matches in the first 2 columns using wildcards
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 2 0 2
11 1 0 4
11 2 0 5
11 3 0 6
------------------------------------------------
Number of Values: 6 Number of Nodes: 2
================================================
Another way to partition results is to use subscripting. You can subscript into the individual imat_result_* objects using logical numeric subscripting, as well as the other input types supported by the partition methods described above. In fact, IMAT uses the partition method directly when you apply subscripting.
Generally, using numeric indices extracts the ith row(s) from the result, as shown in the example below. Currently this subscripting only operates on the underlying data location objects, so to use it with an imat_result you must first extract the data location object.
>> r
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
>> dan=r.data % Extract the Data At Nodes object
dan =
Data At Nodes: 3DOF global translation vector
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 2 0 2
1 3 0 3
------------------------------------------------
Number of Values: 3 Number of Nodes: 1
================================================
>> dan(2) % Extract the 2nd row using numeric subscripting
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 2 0 2
------------------------------------------------
Number of Values: 1 Number of Nodes: 1
================================================
>> dan(dan.dir==3) % Extract the Z direction rows using logical subscripting
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 3 0 3
------------------------------------------------
Number of Values: 1 Number of Nodes: 1
================================================
>> dan('1x') % Extract the 1X result (only valid with Data At Nodes)
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
------------------------------------------------
Number of Values: 1 Number of Nodes: 1
================================================
>> dan(imat_ctrace('1x','1z')) % Extract the 1X and 1Z results using an imat_ctrace
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 3 0 3
------------------------------------------------
Number of Values: 2 Number of Nodes: 1
================================================
To change an attribute of a result, you use the syntax r.attrib=value. Numeric attributes should be set to a numeric array with dimension matching r. String-valued attributes and list attributes should be set to a cell array of strings, and the cell array should be the same dimension as r. There are two special numeric attributes that should have a first extra dimension of 6. These are EffectiveMass and ParticipationFactor.If you set the EffectiveMass, or Participation attribute to a single column vector, then the same vector is used for all elements of r.
A special case is made for times when you want to set all results in an imat_result array to have the same attribute. In this case, you can set the value to be a scalar numeric value or a simple character string. The same value will then be applied to all of the results in the array.
Changing some attributes will affect others. For example, if you change Frequency or ViscousDamping, the Eigenvalue attribute will change.
The set function is an alternative way to change attributes or result data. The syntax is r2=set(r,'attrib',value,...). You can set one or many attributes. The result r2 is a duplicate of r, with the exception of the attributes you specify. set also accepts a structure, like the one that get returns.
Setting the data attribute is straight-forward. Simply set the .data.data property with the new data values, as shown in the example below. The Data attribute should have an extra first dimension as discussed above. If the number of values changes for a given result, the component list will change accordingly.
>> r
r =
2x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- --------------------------------- ------------------------- ---------- ------------
1 DERIVED RESULT Data On Elements At Nodes Structural User defined
2 SUBCASE=2, LOAD ID=2, TYPE=STRESS Data At Nodes On Elements Structural Stress
>> r(1).data.data = [1 2 3 4]';
Setting components can be done one of two ways. The most natural way is to set the .data.component property directly. The second way involves using the setComponents method for each individual DataLocation object. There are reasons why you might choose one method over the other. One reason to set .data.component directly is its ease of use. One potential disadvantage is that when you set the components this way, the data values are set to 0. This is because the component matrix may or may not have the same size as what was there previously, and it may or may not have the same components. Using setComponents allows you to set the data values at the same time as the components. It also lets you specify the components in the internal compressed format, which is discussed in more detail here. Note that internally, IMAT utilizes setComponents when you set the .data.component property directly.
Setting the .data.component matrix directly can be done both at the imat_result level, and individually at the DataLocation object level. In the example below, we will set the data components at the imat_result level by first extracting the components, and then setting them as a subset of the original components. The component matrix will be a multi-dimensional matrix. For example, if your imat_result is a 2x1 vector of Data At Nodes results, and each result has 12 component rows, your component matrix will be 10x3x2. If your imat_result is 1x2, then your component matrix will be 10x3x1x2. If your result contains mixed result types (as shown in the example below), the first 2 dimensions of the component matrix will be the largest of the dimensions of each individual results, and unused rows and columns must contain Nan values.
>> r
r =
2x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- --------------------------------- ------------------------- ---------- ------------
1 DERIVED RESULT Data On Elements At Nodes Structural User defined
2 SUBCASE=2, LOAD ID=2, TYPE=STRESS Data At Nodes On Elements Structural Stress
>> cc = r.data.component % Extract the component matrix, which is 3x6x2 because r is 2x1
cc(:,:,1) =
1 1 1 0 NaN NaN
1 1 2 0 NaN NaN
1 1 3 0 NaN NaN
cc(:,:,2) =
1 1 11 0 0 0
1 1 12 0 0 0
1 1 22 0 0 0
>> r.data.data % Each column contains the data values for that result
ans =
1.23435 6.53175
3.65766 -3.65766
0 0
>> r.data.component = cc(1:2,:,:); % Set the component matrix using the first 2 rows of each
>> r.data.component
cc(:,:,1) =
1 1 1 0 NaN NaN
1 1 2 0 NaN NaN
cc(:,:,2) =
1 1 11 0 0 0
1 1 12 0 0 0
>> r.data.data % Notice that the data values are now 0
ans =
0 0
0 0
>> cc2 = cc(:,1:4,1) % Extract the component submatrix for the first result
cc2 =
1 1 1 0
1 1 2 0
1 1 3 0
>> r(1).data.component = cc2; % Set the component matrix for the first result
Using the setComponents method allows you to set the components and data at the same time. The setComponents method accepts different input arguments for each different DataLocation object type, so please see the help for each.
The DataLocation objects all store the component lists internally in a compressed format if at all possible, as described below. setComponents will automatically attempt to compress component lists that are supplied in an uncompressed form. To store the component lists in compressed form, additional internal attributes are needed. These show up as additional input arguments to the setComponents method for some of the DataLocation types. If you wish to supply components in compressed form to setComponents directly, the getComponents method is a very useful way to see how the individual component columns and related attributes are stored internally. The output of getComponents can be passed directly back in to setComponents.
The example below shows how to create a new Data At Nodes object and populate its components and data. The inputs are supplied in compressed form. The result components consist of directions 1-6 repeated for nodes 1-100.
>> d = imat_result_dan; % Create a new Data At Nodes object
>> d = setComponents(d,1:100,1:6,0,true,1:600); % Supply the components and data in compressed form
>> r(3).data = d; % Place the data into the imat_result object
You can use the subscripting capabilities of the imat_result_* data location objects for individual component and data assignments. Some examples are shown below.
>> r
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
>> dan=r.data
dan =
Data At Nodes: 3DOF global translation vector
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 2 0 2
1 3 0 3
------------------------------------------------
Number of Values: 3 Number of Nodes: 1
================================================
>> dan(2).data = 22
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 2 0 22
1 3 0 3
------------------------------------------------
Number of Values: 3 Number of Nodes: 1
================================================
>> dan(dan.dir==3).seid = 1
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 2 0 22
1 3 1 3
------------------------------------------------
Number of Values: 3 Number of Nodes: 1
================================================
>> dan(imat_ctrace('1x','1z')).node = 2
ans =
Data At Nodes: Unknown
================================================
Node Dir SEID Data
------------------------------------------------
2 1 0 1
1 2 0 22
2 3 1 3
------------------------------------------------
Number of Values: 2 Number of Nodes: 2
================================================
In some cases it is possible to convert results from one DataLocation to another. This process is called typecasting, because you are converting one object type to another.
>> dan = imat_result_dan; % Create an empty Data At Nodes object
>> dan.data=1:12 % Put some data into it
Data At Nodes: 3DOF global translation vector
================================================
Node Dir SEID Data
------------------------------------------------
1 1 0 1
1 2 0 2
1 3 0 3
2 1 0 4
2 2 0 5
2 3 0 6
3 1 0 7
3 2 0 8
3 3 0 9
4 1 0 10
4 2 0 11
4 3 0 12
------------------------------------------------
Number of Values: 12 Number of Nodes: 4
================================================
>> r = imat_result(dan) % Place the data into the imat_result object
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ------------- --------- ------------
1 Data At Nodes Unknown User defined
>> r.data = imat_result_doe(r.data) % Convert the Data At Nodes result into Data On Elements
r =
1x1 IMAT_RESULT
Row Name DataLocation ModelType ResultType
--- ---- ---------------- --------- ------------
1 Data On Elements Unknown User defined
>> r.data % View the typecasted data
Data On Elements
================================================
Element Layer SEID Data
------------------------------------------------
1 1 0 1
1 2 0 2
1 3 0 3
2 1 0 4
2 2 0 5
2 3 0 6
3 1 0 7
3 2 0 8
3 3 0 9
4 1 0 10
4 2 0 11
4 3 0 12
------------------------------------------------
Number of Values: 12 Number of Elements: 4
================================================
Since the component list can be extremely large for a given result, where possible the imat_result class will store the component matrix in a compressed form. If it cannot, it will store them in uncompressed form. When the component list is set or modified, it will always attempt to compress the component list. In addition, the component list is stored internally in separate columns as INT32.
To access the compressed component definition, use getComponents, as shown below. This component definition can be passed in to setComponents directly.
>> c = getComponents(r(1).data)
ans =
[208x1 int32] [816x1 int32] [208x1 double] [6x1 int32] [1616x1 int32] [816x1 int32] [0] [0] [1]
>> r(1).data = setComponents(r(1).data,c{:});
You can perform any MATLAB operation on an imat_result simply by extracting its data values into a numeric array, performing the desired operation, and creating a new imat_result with the resulting data. However, some operations can be performed directly on imat_result objects, using methods built into IMAT. Here is a summary of the available operations:
| Unary Operations | |
| +r | Unary plus (leaves r unchanged) |
| -r | Unary minus (negate the data values of r) |
| real(r) | Take real part of the data values of r |
| imag(r) | Take imaginary part of the data values of r |
| conj(r) | Take complex conjugate of the data values of r |
| abs(r) | Take absolute value of the data values of r (or modulus if r is complex) |
| Binary Operations | |
| r+scalar, r-scalar | Add, subtract scalar value to all data values |
| r.*scalar, r./scalar | Multiply or divide data values by scalar value |
| r+t, r-t | Add or subtract two results. Operation is performed on data values only if data locations and number of values match between r and t. |
| r.*t, r./t | Termwise multiply or divide two shapes. Operation is performed termwise only if nodes of r and t match. |